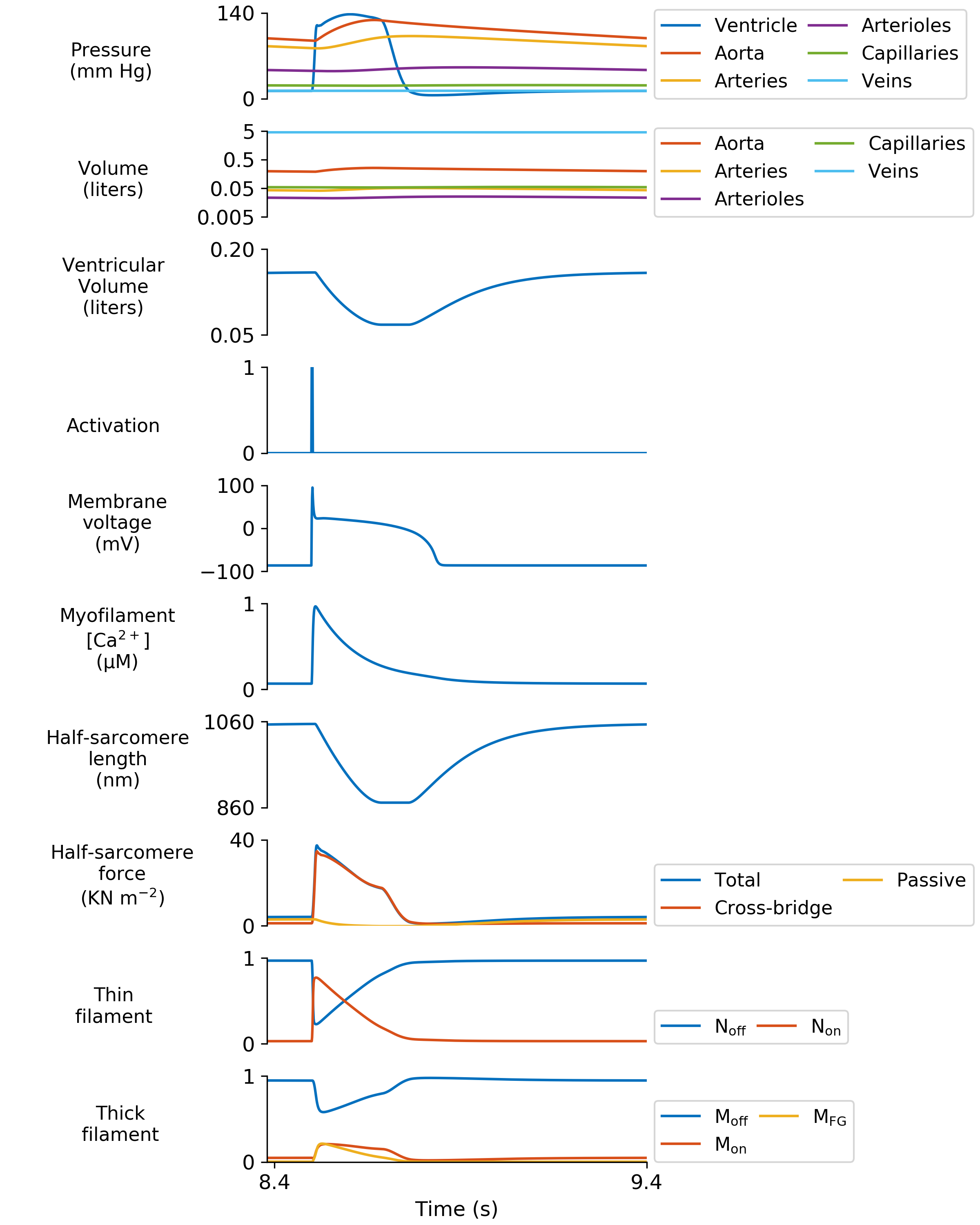
Figure 3
Demos
{
"output_parameters": {
"excel_file": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation.xlsx"],
"csv_file": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation.csv"],
"input_file": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation.json"],
"summary_figure": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation_summary.png"],
"pv_figure": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation_pv.png"],
"baro_figure": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation_baro.png"],
"flows_figure": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation_flows.png"],
"hs_fluxes_figure": ["..\\temp\\frontier_base_simulation\\frontier_base_simulation_hs_fluxes.png"]
},
"baroreflex": {
"baro_scheme": ["fixed_heart_rate"],
"fixed_heart_rate":{
"simulation":{
"no_of_time_points": [17000],
"time_step": [0.001],
"duty_ratio": [0.003],
"basal_heart_period": [1,"s"]
}
},
"simple_baroreceptor":{
"simulation":{
"start_index":[2000],
"memory":[2,"s"],
"no_of_time_points": [150000],
"time_step": [0.001],
"duty_ratio": [0.003],
"basal_heart_period": [1,"s"]
},
"afferent": {
"b_max": [2],
"b_min": [0],
"S": [0.067,"mmHg"],
"P_n": [90,"mmHg"]
},
"regulation":{
"heart_period":{
"G_T": [0.03]
},
"k_1":{
"G_k1": [-0.05]
},
"k_on":{
"G_k_on":[0.02]
},
"ca_uptake":{
"G_up": [-0.02]
},
"g_cal":{
"G_gcal": [-0.03]
}
}
}
},
"perturbations": {
"perturbation_activation":[true],
"volume":{
"start_index": [10000],
"stop_index": [15000],
"increment": [-6e-4]
},
"valve":{
"aortic":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"mitral":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
}
},
"compliance": {
"aorta":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"capillaries": {
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"venous":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
}
},
"resistance": {
"aorta":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"capillaries": {
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"venous":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"ventricle":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
}
},
"myosim":{
"k_1":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"k_2":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"k_4_0":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
}
},
"ca_handling":{
"ca_uptake":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"ca_leak":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
},
"g_cal":{
"start_index": [0],
"stop_index": [0],
"increment": [0]
}
}
},
"circulation":{
"no_of_compartments": [6],
"blood":{
"volume":[5,"liters"]
},
"aorta":{
"resistance": [40,"s"],
"compliance": [0.002,"liter_per_mmHg"]
},
"arteries":{
"resistance": [200,"s"],
"compliance": [0.0005,"liter_per_mmHg"]
},
"arterioles":{
"resistance": [500,"s"],
"compliance": [0.0005,"liter_per_mmHg"]
},
"capillaries":{
"resistance": [300,"s"],
"compliance": [0.0025,"liter_per_mmHg"]
},
"veins":{
"resistance": [100,"s"],
"compliance": [0.35,"liter_per_mmHg"]
},
"ventricle":{
"resistance": [20,"s"],
"wall_volume": [0.1,"liters"],
"slack_volume": [0.08,"liters"],
"wall_density": [1055,"g/l"],
"body_surface_area": [1.90,"m^2"]
}
},
"half_sarcomere":{
"max_rate": [5000,"s^-1"],
"temperature": [288, "Kelvin"],
"cb_number_density": [6.9e16, "number of cb's/m^2"],
"initial_hs_length": [900, "nm"],
"ATPase_activation":[false],
"delta_energy":[70,"kJ/mol"],
"avagadro_number":[6.02e23,"mol^-1"],
"reference_hs_length":[1100,"nm"],
"myofilaments":{
"kinetic_scheme": ["3state_with_SRX"],
"k_1": [2,"s^-1"],
"k_force": [1e-3, "(N^-1)(m^2)"],
"k_2": [200, "s^-1"],
"k_3": [100, "(nm^-1)(s^-1)"],
"k_4_0": [200, "s^-1"],
"k_4_1": [0.1, "nm^-4"],
"k_cb": [0.001, "N*m^-1"],
"x_ps": [5, "nm"],
"k_on": [5e8, "(M^-1)(s^-1)"],
"k_off": [200, "s^-1"],
"k_coop": [5],
"bin_min": [-10, "nm"],
"bin_max": [10, "nm"],
"bin_width": [1, "nm"],
"filament_compliance_factor": [0.5],
"thick_filament_length": [815, "nm"],
"thin_filament_length": [1120, "nm"],
"bare_zone_length": [80, "nm"],
"k_falloff": [0.0024],
"passive_mode": ["exponential"],
"passive_exp_sigma": [500],
"passive_exp_L": [80],
"passive_l_slack": [900, "nm"]
},
"membranes": {
"kinetic_scheme": ["Ten_Tusscher_2004"],
"simple_2_compartment":{
"Ca_content": [1e-3],
"k_leak": [2e-3],
"k_act": [5e-2],
"k_serca": [10.0]
},
"Ten_Tusscher_2004":{
"g_to_factor": [1],
"g_Kr_factor": [1],
"g_Ks_factor": [1],
"Ca_a_rel_factor": [1],
"Ca_V_leak_factor": [1],
"Ca_Vmax_up_factor": [1],
"g_CaL_factor": [1]
}
}
},
"growth": {
"growth_activation": [false],
"start_index": [200000],
"moving_average_window": [5000],
"driven_signal": ["stress"],
"concenrtric":{
"G_stress_driven":[1e-6],
"G_ATPase_driven":[-2]
},
"eccentric":{
"G_number_of_hs":[-3e-6],
"G_ATPase_driven":[0]
}
},
"profiling":{
"profiling_activation":[false]
},
"saving_to_spreadsheet":{
"saving_data_activation":[true],
"output_data_format":["csv"],
"start_index":[0],
"stop_index":[17000]
},
"multi_threads" :{
"multithreading_activation":[false],
"parameters_in":{
"G_wall_thickness": {
"values":[25,50,100,200,300],
"param_out":["ventricle_wall_thickness"],
"section": ["growth"]
},
"G_number_of_hs": {
"values":[25,50,100,200,300],
"param_out":["number_of_hs"],
"section": ["growth"]
}
},
"output_main_folder": ["..\\temp\\frontier_base_simulation\\demo_i_j\\demo_i_j.json"]
}
}
Python Script
def range(x, axis=0):
return np.max(x, axis=axis) - np.min(x, axis=axis)
def display_Fig3 (data_structure, output_file_string="",
t_limits=[],dpi=300):
# unit multipliers
membrane_voltage_factor = 1000 # Convert Voltage to mili-Voltage
myo_ca_transient_factor = 1e6 # Convert Mol to micro-Mol
sarcomeric_force_factor = 1e-3 # Convert N to KN
# y-lim thresholds
pressure_lim = [0,140] # in mm Hg
vol_lim = [0.005,5] # in liters
vent_vol_lim = [0.05,0.20] # in liters
act_lim = [0,1] # unitless
mem_volt_lim = [-100,100] # in mili-Voltage
myof_ca_lim = [0,1] # in micro-Mol
sarc_length_lim = [860,1060] # in nano-meter
sarc_force_lim = [0,40] # in kilo-pascal
thin_fila_lim = [0,1] # unitless
thick_fila_lim = [0,1] # unitless
# Developing plots
no_of_rows = 10
no_of_cols = 1
f = plt.figure(constrained_layout=True)
f.set_size_inches([8, 10])
spec2 = gridspec.GridSpec(nrows=no_of_rows, ncols=no_of_cols,
figure=f,width_ratios=[1])
# ***Plot pressure data***
ax1 = f.add_subplot(spec2[0, 0])
ax1.plot('time', 'pressure_ventricle', data=data_structure, label='Ventricle',color='#0470BE')
ax1.plot('time', 'pressure_aorta', data=data_structure, label='Aorta',color='#D8501A')
ax1.plot('time', 'pressure_arteries', data=data_structure, label='Arteries',color='#EEAF1F')
ax1.plot('time', 'pressure_arterioles', data=data_structure, label='Arterioles',color='#802D90')
ax1.plot('time', 'pressure_capillaries', data=data_structure, label='Capillaries',color='#75AC2F')
ax1.plot('time', 'pressure_veins', data=data_structure, label='Veins',color='#4DBEEF')
# remove the unnecessary ax frames
ax1.spines['top'].set_visible(False)
ax1.spines['right'].set_visible(False)
ax1.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax1.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax1.set_xticks([])
# Y axis
ax1.set_ylim(pressure_lim)
ax1.set_yticks(pressure_lim)
ax1.set_ylabel('Pressure\n(mm Hg)', fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax1.tick_params(labelsize = 12)
ax1.legend(loc=(1.02, 0.0),ncol=2,fontsize = 11,columnspacing=0.6)
# ***Plot Volume data***
ax2 = f.add_subplot(spec2[1, 0])
ax2.semilogy('time', 'volume_aorta', data=data_structure, label='Aorta',color='#D8501A')
ax2.semilogy('time', 'volume_arteries', data=data_structure, label='Arteries',color='#EEAF1F')
ax2.semilogy('time', 'volume_arterioles', data=data_structure, label='Arterioles',color='#802D90')
ax2.semilogy('time', 'volume_capillaries', data=data_structure, label='Capillaries',color='#75AC2F')
ax2.semilogy('time', 'volume_veins', data=data_structure, label='Veins',color='#4DBEEF')
# remove the unnecessary ax frames
ax2.spines['top'].set_visible(False)
ax2.spines['right'].set_visible(False)
ax2.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax2.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax2.set_xticks([])
# Y_axis
ax2.set_ylim(vol_lim)
ax2.minorticks_off()
y_ticks = [0.005,0.05,0.5, 5]
ax2.set_yticks(y_ticks)
ax2.get_yaxis().set_major_formatter(tk.FixedFormatter(seq =y_ticks))
ax2.set_ylabel('Volume\n(liters)', fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax2.tick_params(labelsize = 12)
ax2.legend(loc=(1.02, 0.0),fontsize = 11, ncol=2,columnspacing=0.6)
# ***Plot vetricular volume data***
ax3 = f.add_subplot(spec2[2, 0])
ax3.plot('time', 'volume_ventricle', data=data_structure, label='Ventricle',color='#0470BE')
# remove the unnecessary ax frames
ax3.spines['top'].set_visible(False)
ax3.spines['right'].set_visible(False)
ax3.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax3.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax3.set_xticks([])
# Y_axis
ax3.set_ylim(vent_vol_lim)
ax3.set_yticks(vent_vol_lim)
ax3.set_ylabel('Ventricular\nVolume\n(liters)', fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax3.tick_params(labelsize = 12)
# ***Plot activation data***
ax4 = f.add_subplot(spec2[3, 0])
ax4.plot('time', 'activation', data=data_structure, label='Activation',color='#0470BE')
# remove the unnecessary ax frames
ax4.spines['top'].set_visible(False)
ax4.spines['right'].set_visible(False)
ax4.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax4.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax4.set_xticks([])
#Y_axis
ax4.set_ylim(act_lim)
ax4.set_yticks(act_lim)
ax4.set_ylabel('Activation', fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax4.tick_params(labelsize = 12)
# ***Plot membrane coltage data***
ax5 = f.add_subplot(spec2[4, 0])
# convert the y_axis from voltage to mili_voltage
y_axis = data_structure['membrane_voltage']*membrane_voltage_factor
x_axis = data_structure['time']
ax5.plot(x_axis, y_axis, label='Voltage',color='#0470BE')
# remove the unnecessary ax frames
ax5.spines['top'].set_visible(False)
ax5.spines['right'].set_visible(False)
ax5.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax5.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax5.set_xticks([])
# Y_axis
ax5.set_ylim(mem_volt_lim)
ax5.set_yticks([mem_volt_lim[0],0,mem_volt_lim[1]])
ax5.set_ylabel('Membrane\nvoltage\n(mV)', fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax5.tick_params(labelsize = 12)
# ***Plot myofilament Ca transient data***
ax6 = f.add_subplot(spec2[5, 0])
# convert the y_axis from Mol to micro-Mol
y_axis = data_structure['Ca_conc']*myo_ca_transient_factor
x_axis = data_structure['time']
ax6.plot(x_axis, y_axis, label='Ca concentration',color='#0470BE')
# remove the unnecessary ax frames
ax6.spines['top'].set_visible(False)
ax6.spines['right'].set_visible(False)
ax6.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax6.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax6.set_xticks([])
# Y_axis
ax6.set_ylim(myof_ca_lim)
ax6.set_yticks(myof_ca_lim)
ax6.set_ylabel('Myofilament\n$[Ca^{2+}]$\n(\u03BCM)' , fontsize = 11,rotation=0,labelpad=20,y=0.2)
# other adjustments
ax6.tick_params(labelsize = 12)
# ***Plot half-sarcomere length data***
ax7 = f.add_subplot(spec2[6, 0])
ax7.plot('time', 'hs_length', data=data_structure, label='hs_length',color='#0470BE')
# remove the unnecessary ax frames
ax7.spines['top'].set_visible(False)
ax7.spines['right'].set_visible(False)
ax7.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax7.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax7.set_xticks([])
# Y_axis
ax7.set_ylim(sarc_length_lim)
ax7.set_yticks(sarc_length_lim)
ax7.set_ylabel('Half-sarcomere\nlength\n(nm)', fontsize = 11,rotation=0,labelpad=35,y=0.2)
# other adjustments
ax7.tick_params(labelsize = 12)
# ***Plot half-sarcomere force data***
ax8 = f.add_subplot(spec2[7, 0])
# convert the y_axis from pascal to kilo-pascal
y1_axis = data_structure['hs_force']*sarcomeric_force_factor
y2_axis = data_structure['cb_force']*sarcomeric_force_factor
y3_axis = data_structure['pas_force']*sarcomeric_force_factor
x_axis = data_structure['time']
ax8.plot(x_axis, y1_axis,label='Total',color='#0470BE')
ax8.plot(x_axis, y2_axis,label='Cross-bridge',color='#D8501A')
ax8.plot(x_axis, y3_axis,label='Passive',color='#EEAF1F')
# remove the unnecessary ax frames
ax8.spines['top'].set_visible(False)
ax8.spines['right'].set_visible(False)
ax8.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax8.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax8.set_xticks([])
# Y_axis
ax8.set_ylim(sarc_force_lim)
ax8.set_yticks(sarc_force_lim)
ax8.set_ylabel('Half-sarcomere\nforce\n(KN $m^{-2}$)', fontsize = 11,rotation=0,labelpad=40,y=0.2)
# other adjustments
ax8.tick_params(labelsize = 12)
ax8.legend(loc=(1.02, 0.0),fontsize = 11,ncol=2,columnspacing=0.6)
# ***Plot thin-filament data***
ax9 = f.add_subplot(spec2[8, 0])
ax9.plot('time', 'n_off', data=data_structure, label='$N_{off}$',color='#0470BE')
ax9.plot('time', 'n_on', data=data_structure, label='$N_{on}$',color='#D8501A')
# remove the unnecessary ax frames
ax9.spines['top'].set_visible(False)
ax9.spines['right'].set_visible(False)
ax9.spines['bottom'].set_visible(False)
# X_axis
if t_limits:
ax9.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax9.set_xticks([])
# Y_axis
ax9.set_ylim(thin_fila_lim)
ax9.set_yticks(thin_fila_lim)
ax9.set_ylabel('Thin\nfilament', fontsize = 11,rotation=0,labelpad=45,y=0.2)
# other adjustments
ax9.tick_params(labelsize = 12)
ax9.legend(loc=(1.02, 0.0),fontsize = 11,ncol=2,columnspacing=0.6)
# ***Plot thick-filament data***
ax10 = f.add_subplot(spec2[9, 0])
ax10.plot('time', 'M_OFF', data=data_structure, label='$M_{off}$',color='#0470BE')
ax10.plot('time', 'M_ON', data=data_structure, label='$M_{on}$',color='#D8501A')
ax10.plot('time', 'M_bound', data=data_structure, label='$M_{FG}$',color='#EEAF1F')
# remove the unnecessary ax frames
ax10.spines['top'].set_visible(False)
ax10.spines['right'].set_visible(False)
# X_axis
if t_limits:
ax10.set_xlim([t_limits[0]-0.02*range(t_limits),t_limits[1]])
ax10.set_xticks(t_limits)
ax10.set_xlabel('Time (s)',fontsize=12)
# Y_axis
ax10.set_ylim(thick_fila_lim)
ax10.set_yticks(thick_fila_lim)
ax10.set_ylabel('Thick\nfilament', fontsize = 11,rotation=0,labelpad=45,y=0.2)
# other adjustments
ax10.tick_params(labelsize = 12)
ax10.legend(loc=(1.02, 0.0),fontsize = 11,ncol=2,columnspacing=0.6)
# change the mathematical notatio format
rcParams['mathtext.default'] = 'regular'
f.align_ylabels()
# save the figure
if (output_file_string):
save_figure_to_file(f, output_file_string, dpi)
def save_figure_to_file(f, im_file_string, dpi=None, verbose=0):
# Writes an image to file
import os
from skimage.io import imsave
# Check directory exists and save image file
dir_path = os.path.dirname(im_file_string)
if not os.path.isdir(dir_path):
os.makedirs(dir_path)
if (verbose):
print('Saving figure to to %s' % im_file_string)
f.savefig(im_file_string, dpi=dpi)
if __name__ == "__main__":
# import required libraries
import os
import sys
import json
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import matplotlib.ticker as tk
from matplotlib import rcParams
no_of_arguments = len(sys.argv)
if (no_of_arguments == 1):
print('Analysis called with no inputs')
elif (no_of_arguments == 2):
print('Running model %s' % sys.argv[1])
data_path = sys.argv[1]
data = pd.read_csv(data_path)
current_directory = os.getcwd()
display_Fig3(data,current_directory+'/'+'Fig_3.png',[8.4,9.4])
Output Figure