Organization
This software gives you the flexibility to analyze experimental designs that have n-factors.
For example, you may be testing how force-pCa curves vary in myocardium from the left and right ventricles in patients who do or do not have heart failure. In this case, you have a two-factor design.
factor 1is ventricle and can take the values of- left
- right
factor_2is heart_failure_status and can take the values of- heart_failure
- non_failing
If you run different experiments where you only compare cells from patients with diabetes to cells from patients without diabetes, you have a single factor design. In this case:
factor_1is diabetic_status and can take values of- yes
- no
It is also likely that you will have data from multiple preparations from several different people or animals. For example, you might have 2, 3, or 4 cells from each of 12 people. The source of the cells - that is whether they came from John Doe or Jane Doe - is important because it can be used as a grouping variable to increase statistical power.
We call the source of the cells the tag. If you are working with tissue from our biobank, the tag is the patients’s hashcode (for example, 117DD). If you are working with data from animals, you could use an identity number for the mouse or simply write something like Mouse_1.
The last organizational level is the preparation itself - the specific cell or trabecula that you were analyzing. In most cases, you will have recorded multiple SLControl files from a single preparation. All of the files, and nothing else, should go in a single folder that has a unique name. The name of the folder identifies the prep.
The recomended approach is to name the folder with the data followed by ‘a’, ‘b’, ‘c’, etc. to indicate whether it is the first, second, or third prep you analyzed from the tag on that day. For example, 29May2020a, 29May2020b, 29May2020c.
Putting all this together, you should organize your SLControl files in a folder structure of the form
<project>/raw_data/factor_1/factor_2/tag/prep
Tip
- Avoid using spaces in folder and file names.
- For example, use
Heart_failureas opposed toHeart failure
- For example, use
Example
Here is an example layout
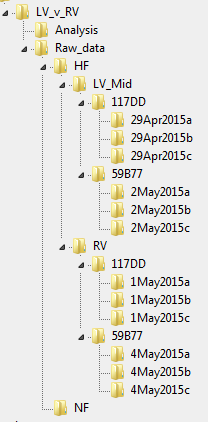
Explanation
Here’s the explanation.
- Everything do with this mechanics experiments is in the project folder called LV_v_RV
- All of the SLControl data is in a single folder called Raw_data. Nothing else ever goes in that folder.
- Raw_data is divided into 2 folders, HF and NF. These are the groups that form Factor 1.
- The HF folder contains 2 folders, LV_Mid and RV. These are the groups that form Factor 2.
- The LV_Mid folder contains two folders, 117DD and 59B77. These are two tags.
- The RV folder contains the same two folders, 117DD and 59B77, because we are analyzing samples from different regions of the same heart.
- Each tag folder contains 3 prep folders.
In summary, we analyzed 12 preparations from the LV or the RV of 2 separate people.
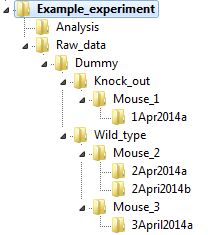
Single factor experiments
In the example below, factor 1 (knock_out or wild_type) is inside a single folder called Dummy.