Pendulum
Overview
This demo builds on the pendulum_1 demo and shows a pendulum connected to a muscle that is transiently activated.
What this demo does
This demo runs a single simulation with a pendulum
- released from its zero position
- connected to a MATMyoSim model that
- has passive stiffness
- is transiently activated by bursts of Ca2+
Instructions
- In MATLAB, change the working directory to
<repo>/code/demos/pendulum/pendulum_2 - open
demo_pendulum_2.m - Press F5 to run
Output
You should see two figures
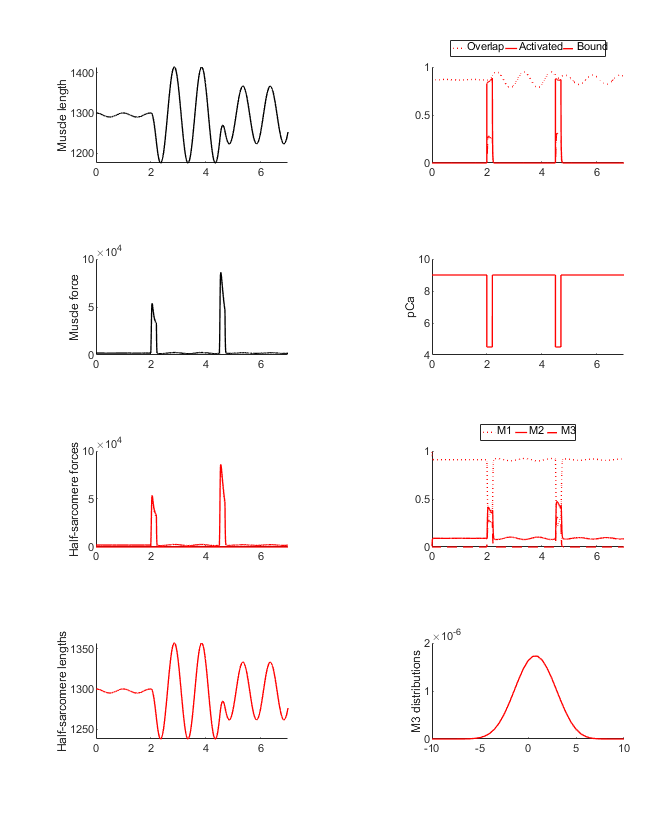
and
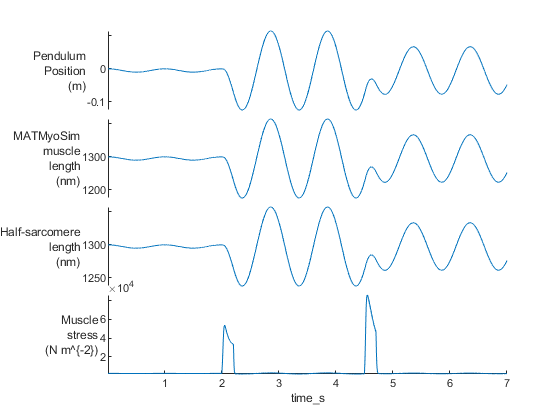
How this worked
This demo is very similar to pendulum_1 demo except that the pendulum is started with an initial position of 0 and an initial velocity of 0. These are defined in the last entry in sim_input/pendulum.json.
{
"pendulum":
{
"m": 10,
"L": 0.25,
"g": 9.81,
"eta": 0,
"hsl_scaling_factor": 500,
"force_scaling_factor": 1e-3,
"initial_conditions": [0.0, 0]
}
}
The other key difference is that the MATMyoSim muscle is transiently activated by 2 bursts of Ca2+. These are defined in the demo_pendulum_2.m file.
% Brief activations
pCa(2000:2200) = 4.5;
pCa(4500:4700) = 4.5;
The first pulse generates a transient force that increases the magnitude of the pendulum’s swing. The second pulse is timed to partially brake the motion.
The first section of the demo code is
function demo_pendulum_1
% Variables
model_file = 'sim_input/model.json'
pendulum_file = 'sim_input/pendulum.json'
options_file = 'sim_input/options.json'
output_file = 'sim_output/output.myo'
% Make sure the path allows us to find the right files
addpath(genpath('../../../../code'));
% Create a simulation
sim = simulation(model_file)
% Set up a mini protocol with the number of time-points
% and pCa held at 9
no_of_time_points = 5000;
pCa = 9.0 * ones(no_of_time_points, 1);
dt = 0.001*ones(no_of_time_points,1);
This sets some filenames, updates the MATLAB path, and initialises a simulation based on a MATMyoSim model file.
The last three lines create a mini protocol with:
- 5000 time-points
- each with a pCa value of 9.0
- and a time-step of 0.001 s
The next section
% Implement the protocol
sim.implement_pendulum_protocol( ...
'pendulum_file_string', pendulum_file, ...
'options_file_string', options_file, ...
'dt', dt, ...
'pCa', pCa);
% Save the output
sim_output = sim.sim_output;
if (~isempty(output_file))
% Check directory exists
output_dir = fileparts(output_file);
if (~isdir(output_dir))
sprintf('Creating output directory: %s', fullfile(cd,output_dir))
[status, msg, msgID] = mkdir(output_dir);
end
save(output_file,'sim_output');
end
implements the pendulum protocol and saves the output to a MATLAB file.
The last section
% Convert sim_output to a table - this requires deleting some fields
sim_output = sim.sim_output;
sim_output = rmfield(sim_output, 'myosim_muscle');
sim_output = rmfield(sim_output, 'subplots');
sim_output = rmfield(sim_output, 'no_of_time_points');
sim_output = rmfield(sim_output, 'cb_pops')
sim_output = struct2table(sim_output);
% Draw some of the output fields as a stacked plot
figure(3);
clf
stackedplot(sim_output, ...
{'pendulum_position', 'muscle_length', 'hs_length', ...
'hs_force'}, ...
'XVariable', 'time_s', ...
'DisplayLabels', { ...
{'Pendulum','Position','(m)'}, ...
{'MATMyoSim','muscle','length','(nm)'}, ...
{'Half-sarcomere','length','(nm)'}, ...
{'Muscle','stress','(N m^{-2})'}});
converts the output data to a MATLAB table and plots some of the key variables using the stackedplot function.
Interpretation
The sim_input/pendulum.json file defined the initial conditions of the pendulum as a position of 0.05 m and 0 velocity.
{
"pendulum":
{
"m": 10,
"L": 0.25,
"g": 9.81,
"eta": 0,
"hsl_scaling_factor": 500,
"force_scaling_factor": 1e-3,
"initial_conditions": [0.05, 0]
}
}
The activating Ca2+ concentration remained low throughout the simulation (pCa = 9.0 throughout) so the pendulum swung perturbed only by the passive stiffness of the MATMyoSim muscle.