Ramp 3 demonstration
This demo shows how to run a simulation of a single sarcomere subjected to a ramp, hold, and then release protocol.
It builds on ramp_1 by adding in a population of cross-bridges that cycle but do not generate force.
Instructions
- Launch MATLAB.
- Change directory to the
MATMyoSim\code\demos\ramps\ramp_3folder in MATLAB. - Open
demo_ramp_3.m. - Press F5 to run the demo.
Code
function demo_ramp_3
% Function illustrates how to run a simulation of a single-half-sarcomere
% with a linear passive elastic component and cycling cross-bridges
% that do not generate active force
% Variables
protocol_file_string = 'ramp_3_protocol.txt';
model_parameters_json_file_string = 'ramp_3_parameters.json';
options_file_string = 'ramp_3_options.json';
model_output_file_string = '..\..\temp\ramp_3_output.myo';
% Make sure the path allows us to find the right files
addpath(genpath('..\..\..\..\code'));
% Run a simulation
sim_output = simulation_driver( ...
'simulation_protocol_file_string', protocol_file_string, ...
'model_json_file_string', model_parameters_json_file_string, ...
'options_json_file_string', options_file_string, ...
'output_file_string', model_output_file_string);
% Load it back up and display to show how that can be done
sim = load(model_output_file_string,'-mat')
sim_output = sim.sim_output
figure(2);
clf;
subplot(2,1,1);
plot(sim_output.time_s,sim_output.muscle_force,'b-');
ylabel('Force (N m^{-2})');
subplot(2,1,2);
plot(sim_output.time_s,sim_output.hs_length,'b-');
ylabel('Half-sarcomere length (nm)');
xlabel('Time (s)');
Model
The model is defined in repo\code\demos\ramps\ramp_3\ramp_3_parameters.json.
{
"MyoSim_model":
{
"muscle_props":
{
"no_of_half_sarcomeres": 1,
"series_k_linear": 0
},
"hs_props":
{
"kinetic_scheme": "3state_with_SRX",
"hs_length": 1300,
"myofilaments":
{
"bin_min": -10,
"bin_max": 10,
"bin_width": 0.5,
"thick_filament_length": 815,
"thin_filament_length": 1120,
"bare_zone_length": 80,
"k_falloff": 0
},
"parameters":
{
"k_1": 10,
"k_force": 0.0,
"k_2": 100,
"k_3": 1,
"k_4_0": 20,
"k_4_1": 0.1,
"k_cb": 0.001,
"x_ps": 0,
"k_on": 6e7,
"k_off": 200,
"k_coop": 10,
"passive_force_mode": "linear",
"passive_hsl_slack": 1250,
"passive_k_linear": 100,
"compliance_factor": 0.5,
"cb_number_density": 6.9e16,
"k_boltzmann": 1.38e-23,
"temperature": 288,
"max_rate": 5000
}
}
}
}
Output
Simulation output
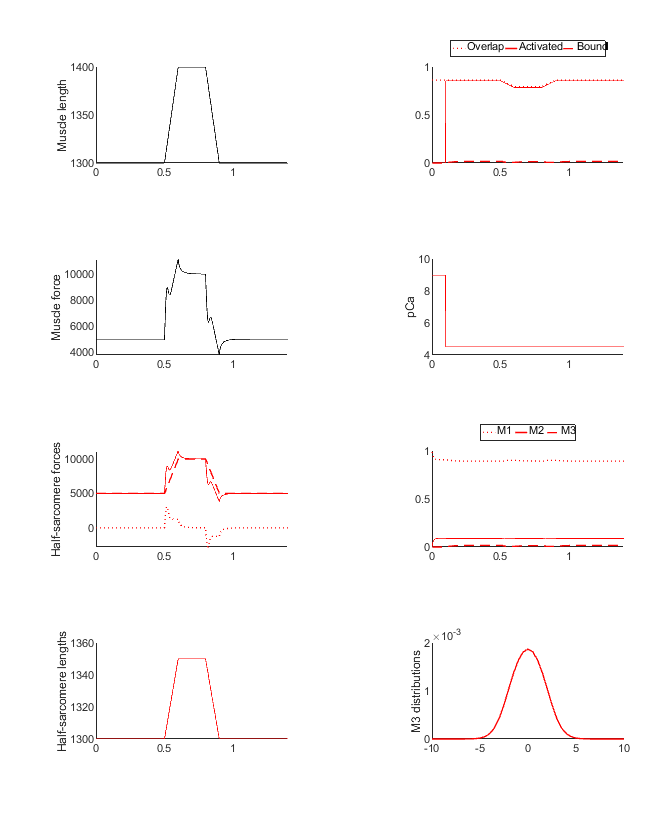
Replotted from output file 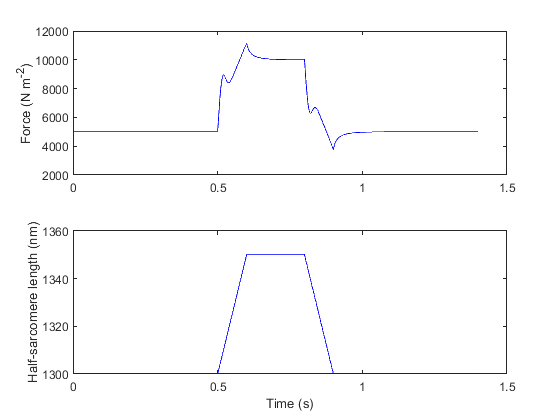
Comments
If you look at the json model structure, you will see that
- the passive component is 10 times stiffer than it was in ramp_1
- passive_k_linear is 100 N m-2 nm-1 instead of 10 N m-2 nm-1)
- k_3 = 1 s-1 nm-1
- As shown in 3state_with_SRX this allows myosin heads to attach to the thin filament
- x_ps = 0 nm
- Thus the attached cross-bridges do not generate active force. They do however link the filaments. If the filaments move relative to each other, the attached cross-bridges generate a drag.
The force produced by this model is thus the sum of the linear passive elastic force and a drag from attached cross-bridges. The cycling kinetics of the heads mean that the cross-bridges produce a visco-elastic drag.