Myosin
Thick filaments are composed of myosin and myosin-binding protein C. The kinetics scheme for both molecules is defined in the model file.
The following test verifies myosin kinetics for the 3-state model schematized below.
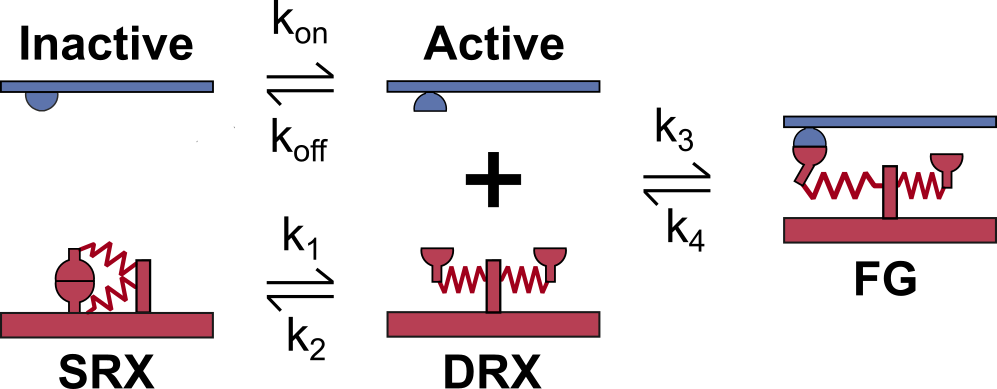
In this model, dimers of myosin heads (red) can transition together from the super-relaxed state (SRX) to the disordered-relaxed state (DRX). Then each myosin head is able to bind to an active binding site (blue) and transition to a force-generating state (FG).
What this test does
The myosin kinetics test:
-
Runs a simulations in which a half-sarcomere is held isometric and activated in a solution with a pCa of 4.5.
-
Saves a status file at each time step, which contains the state of every myosin molecules.
-
Assesses all the myosin transitions occuring between two consecutive time-steps, and calculates the apparent rate constants.
-
Compares the calculated rate constants with those provided in the following model file (the transition parameters between each states are described in the
transitionblocks):
"m_kinetics": [
{
"no_of_states": 3,
"max_no_of_transitions": 2,
"scheme": [
{
"number": 1,
"type": "S",
"extension": 0,
"transition": [
{
"new_state": 2,
"rate_type": "force_dependent",
"rate_parameters": [
100,
100
]
}
]
},
{
"number": 2,
"type": "D",
"extension": 0,
"transition": [
{
"new_state": 1,
"rate_type": "constant",
"rate_parameters": [
200
]
},
{
"new_state": 3,
"rate_type": "gaussian",
"rate_parameters": [
200
]
}
]
},
{
"number": 3,
"type": "A",
"extension": 7.0,
"transition": [
{
"new_state": 2,
"rate_type": "poly",
"rate_parameters": [
150,
1,
2
]
}
]
}
]
}
]
Instructions
Before proceeding, make sure that you have followed the installation instructions and that you already tried to run the single trials demos..
Getting ready
-
Open an Anaconda Prompt
- Activate the FiberSim Anaconda Environment by executing:
conda activate fibersim - Change directory to
<FiberSim_dir>/code/FiberPy/FiberPy, where<FiberSim_dir>is the directory where you installed FiberSim.
Run the test
- Type:
python Fiberpy.py run_batch "../../../testing_suite/thick_kinetics/myosin/batch_m_kinetics.json" - You should see text appearing in the terminal window, showing that the simulations are running. When it finishes (this can take ~20-30 minutes), you should see something similar to the image below.
Viewing the results
The results and summary figure from the simulation are written to files in <FiberSim_dir>/testing_suite/thick_kinetics/myosin/sim_output
The hs folder contains the status files that were dumped at each time-step calculation.
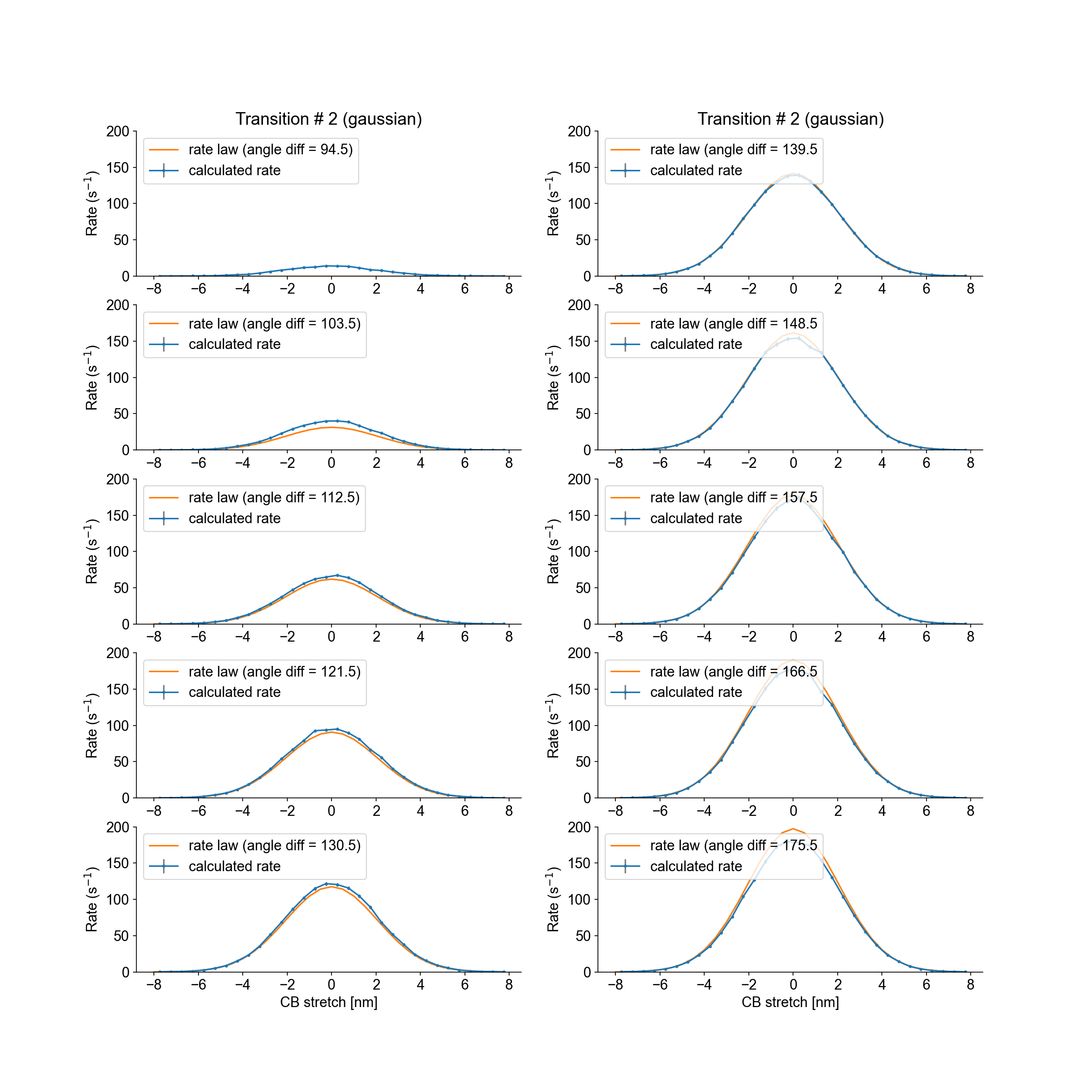
The analysis folder contains figure files showing the calculated rate laws for all myosin transitions, as well as the model rate laws.
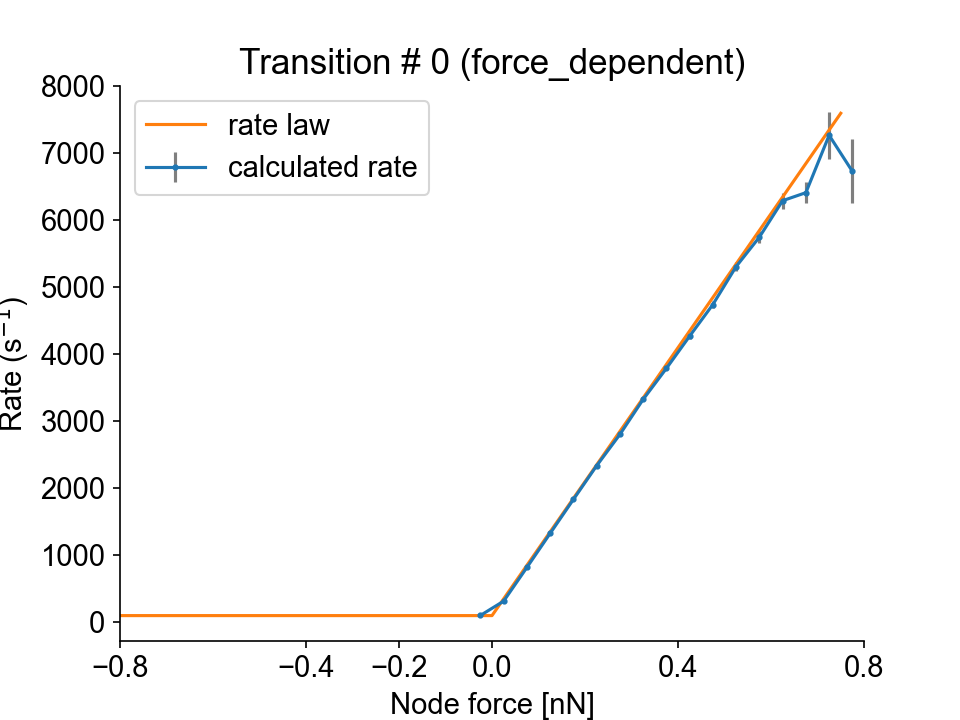
Transition #0 (from state 1 to state 2) is force-dependent, and the transition rate is plotted as a function of the node force.
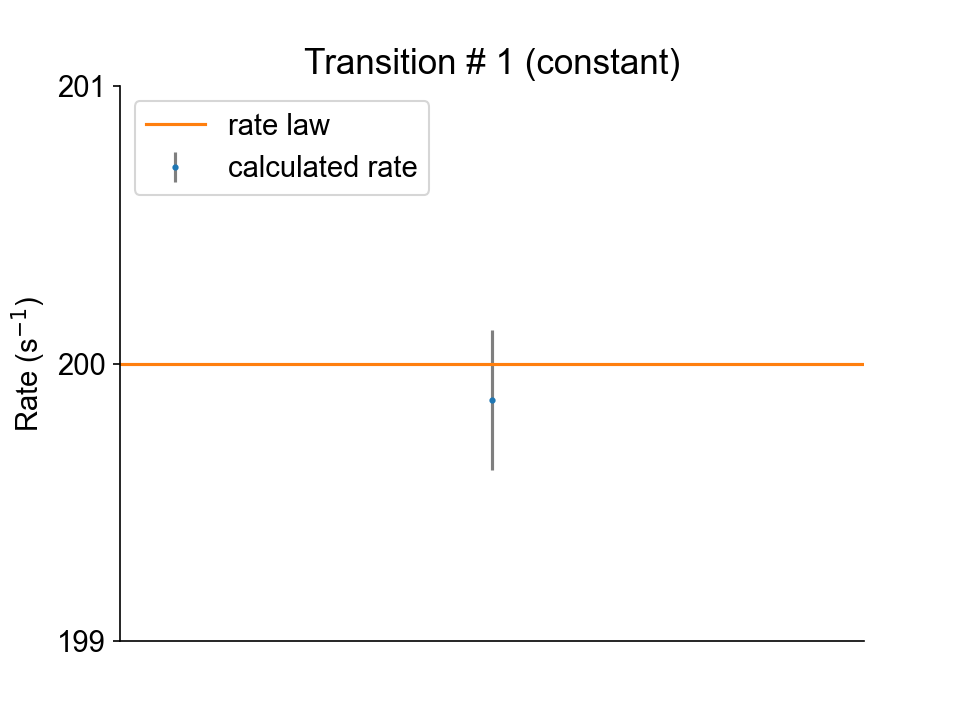
Transition #1 (from state 2 to state 1) is constant.
Transition #2 (from state 2 to state 3) is stretch-dependent, and the transition rate is plotted as a function of the stretch values. Furthermore, the attachement probabilities are modulated by the angle difference between a myosin head and the nearest binding sites. Each panel represents an angle difference interval and shows the calculated and theoretical attachment rates for the mid-interval angle difference.
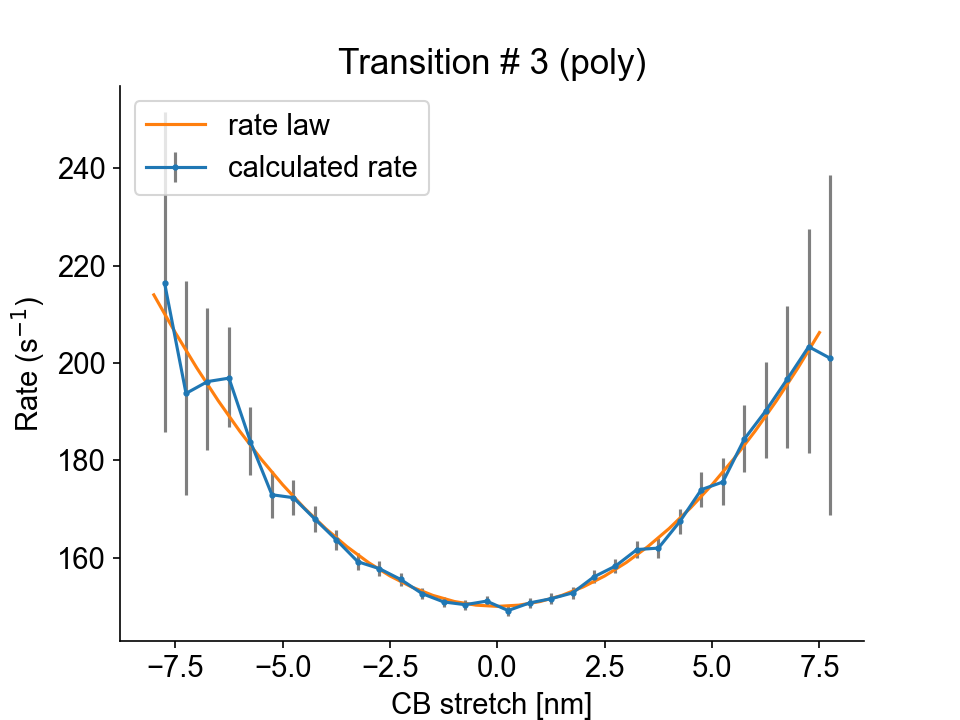
Transition #3 (from state 3 to state 2) is stretch-dependent, and the transition rate is plotted as a function of the stretch values.