Fitting to force-pCa and k_tr
Overview
This demo shows how to fit a model to force-pCa and k_tr data.
What this demo does
This demo:
- uses a fitting setup to iteratively adjust a
base modelseeking the best fit totarget data. - shows how to analyze the fitting process.
Instructions
If you need help with these step, check the installation instructions.
- Open an Anaconda prompt
- Activate the FiberSim environment
- Change directory to
<FiberSim_repo>/code/FiberPy/FiberPy - Run the command
Note before you run this command that initializing a new fit will over-write the example data stored in your repo. Regenerating it will take a long time - potentially several hours. You may want to read through the rest of this page first.
python FiberPy.py fit_model "../../../demo_files/fitting/force_pCa_and_k_tr/base/setup.json"
- You should see text appearing in the terminal window, showing that the simulations are running. When it finishes (this may take a few minutes), you should see something similar to the image below.
Viewing the results
All of the results from the simulation are written to files in <FiberSim_repo>/demo_files/fitting/force_pCa_and_k_tr
Lots of stuff here.
How this worked
The setup file is shown below.
Most parts are standard.
- the
FiberCpp_exesection is as normal. - the
modelsection was described in fitting - the
characterizationsection defines a force-pCa with k_tr protocol.
{
"FiberSim_setup":
{
"FiberCpp_exe": {
"relative_to": "this_file",
"exe_file": "../../../../bin/FiberCpp.exe"
},
"model":
{
"relative_to": "this_file",
"options_file": "sim_options.json",
"fitting":
{
"base_model": "model.json",
"generated_folder": "../generated",
"working_folder": "../working",
"progress_folder": "../progress",
"Python_objective_call": "../Python_code/return_fit.py",
"optimizer": "particle_swarm",
"initial_guess": [0.5, 0.5, 0.5],
"single_run": "False",
"adjustments":
[
{
"variable": "m_kinetics",
"isotype": 1,
"state": 2,
"transition": 1,
"parameter_number": 1,
"factor_bounds": [-1, 1],
"factor_mode": "log"
},
{
"variable": "m_kinetics",
"isotype": 1,
"state": 3,
"transition": 1,
"parameter_number": 1,
"factor_bounds": [-1, 1],
"factor_mode": "log"
},
{
"class": "thin_parameters",
"variable": "a_k_on",
"output_type": "float",
"factor_bounds": [0.5, 1.5]
}
]
}
},
"characterization":
[
{
"type": "pCa_length_control",
"relative_to": "this_file",
"sim_folder": "../sim_data",
"m_n": 25,
"pCa_values": [9, 6.1, 5.8, 5.6, 4.5],
"sim_duration_s": 1.4,
"time_step_s": 0.001,
"pCa_step_up_s": 0.05,
"k_tr_start_s": 0.8,
"k_tr_duration_s": 0.02,
"k_tr_ramp_s": 0.001,
"k_tr_magnitude_nm": 100,
"k_tr_fit_time_s": [0.822, 1.39],
"output_image_formats": [ "png" ],
"figures_only": "False",
"trace_figures_on": "False"
}
]
}
}
The only new feature included is the Python_objective_call. This defines a relative path to a Python script reproduced below.
The script:
- accesses the results of a test simulation and extracts values of:
- Fmax
- Fmin
- pCa50
- nH
- value of ktr calculated at:
- pCa 4.5
- pCa 5.6
- pCa 5.8
- compares these values to corresponding data stored in
../target_data/target_data.xlsx - updates several files with information about the fitting process
- returns a single value quantifying the least-squares error between the test simulation and the
target_data.
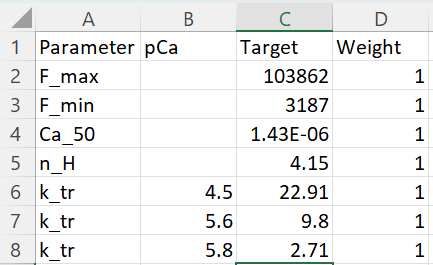
The target_data file is shown here.
Python script
# -*- coding: utf-8 -*-
"""
Created on Mon Jan 22 21:56:54 2024
@author: ken
"""
import os
import sys
from pathlib import Path
import numpy as np
import pandas as pd
def return_fit():
""" returns a single value defining the least squares fit
between the current simulation and the target data """
# Variables
top_data_folder = '../sim_data/sim_output'
pCa_data_file = 'pCa_analysis.xlsx'
pCa_sheet = 'curve_1'
k_tr_data_file = 'k_tr_analysis.xlsx'
target_data_file = '../target/target_data.xlsx'
trial_errors_file = '../working/trial_errors.xlsx'
comparison_file = '../sim_data/sim_output/comparison.xlsx'
# Code
# Adapt because files are relative to this file
parent_dir = Path(__file__).parent.absolute()
top_data_folder = Path(os.path.join(parent_dir, top_data_folder)).resolve()
# Open the pCa data
pCa_data_file = os.path.join(top_data_folder, pCa_data_file)
pCa_data = pd.read_excel(pCa_data_file,
sheet_name = pCa_sheet)
# Open the k_tr file
k_tr_data_file = os.path.join(top_data_folder, k_tr_data_file)
k_tr_data = pd.read_excel(k_tr_data_file)
# Open the target_data
target_data_file = os.path.join(parent_dir, target_data_file)
target_data = pd.read_excel(target_data_file)
# Set the trial errors file
trial_errors_file = os.path.join(parent_dir, trial_errors_file)
# Set the comparison file
comparison_file = os.path.join(parent_dir, comparison_file)
# Display
print(pCa_data)
print(k_tr_data)
print(target_data)
# Build up the error signal
no_of_error_components = 7
error_components = np.NaN * np.ones(no_of_error_components)
# Cycle through them
for i in range(no_of_error_components):
# Pull test values from simulation spreadsheets
if (i==0):
test_value = pCa_data['y_min'].iloc[0] + \
pCa_data['y_amp'].iloc[0]
# Add in a column for test values
target_data['Test_value'] = np.NaN * np.ones(len(target_data['Target']))
if (i==1):
test_value = pCa_data['y_min'].iloc[0]
target_value = target_data['Target'].iloc[1]
w = target_data['Weight'].iloc[1]
if (i==2):
test_pCa = pCa_data['pCa_50'].iloc[0]
test_value = np.power(10, -test_pCa)
if (i==3):
test_value = pCa_data['n_H'].iloc[0]
if (i==4):
test_value = k_tr_data['k_tr'].iloc[0]
if (i==5):
test_value = k_tr_data['k_tr'].iloc[1]
if (i==6):
test_value = k_tr_data['k_tr'].iloc[2]
# Pull targets
target_value = target_data['Target'].iloc[i]
w = target_data['Weight'].iloc[i]
# Add test_value to target
target_data.loc[i, 'Test_value'] = test_value
# Calculate error_component
error_components[i] = w * np.power(
(target_value - test_value) / target_value, 2.0)
# Calculate final value
e = np.sum(error_components)
# Write error data to file
d = dict()
for i in range(len(error_components)):
d['error_cpt_%i' % (i+1)] = error_components[i]
d['error_total'] = e
# Make a dataframe
df = pd.DataFrame(data=d, index=[0])
print(target_data)
print(df)
# Check the dir exists
worker_parent_dir = Path(trial_errors_file).parent
if not os.path.isdir(worker_parent_dir):
os.makedirs(worker_parent_dir)
# Clean the file and then write
if (os.path.exists(trial_errors_file)):
os.remove(trial_errors_file)
df.to_excel(trial_errors_file, index=False)
# Now write the test_values file
target_data.to_excel(comparison_file, index=False)
if __name__ == "__main__":
return_fit()