Half-sarcomeres with variable properties
Overview
This demo shows how to simulate multiple half-sarcomeres that have variable properties.
What this demo does
This demo:
- Builds on the multiple half-sarcomeres with series compliance demo but reduces the attachment rate for myosin in a half-sarcomere in the middle of the myofibril
- Plots summaries of the simulation
Instructions
If you need help with these step, check the installation instructions.
- Open an Anaconda prompt
- Activate the FiberSim environment
- Change directory to
<FiberSim_repo>/code/FiberPy/FiberPy - Run the command
python FiberPy.py characterize "../../../demo_files/myofibrils/variable_hs/base/setup.json" - You should see text appearing in the terminal window, showing that the simulations are running. It may take a few minutes to finish.
Viewing the results
All of the results from the simulation are written to files in <FiberSim_repo>/demo_files/myofibrils/variable_hs/sim_data/sim_output
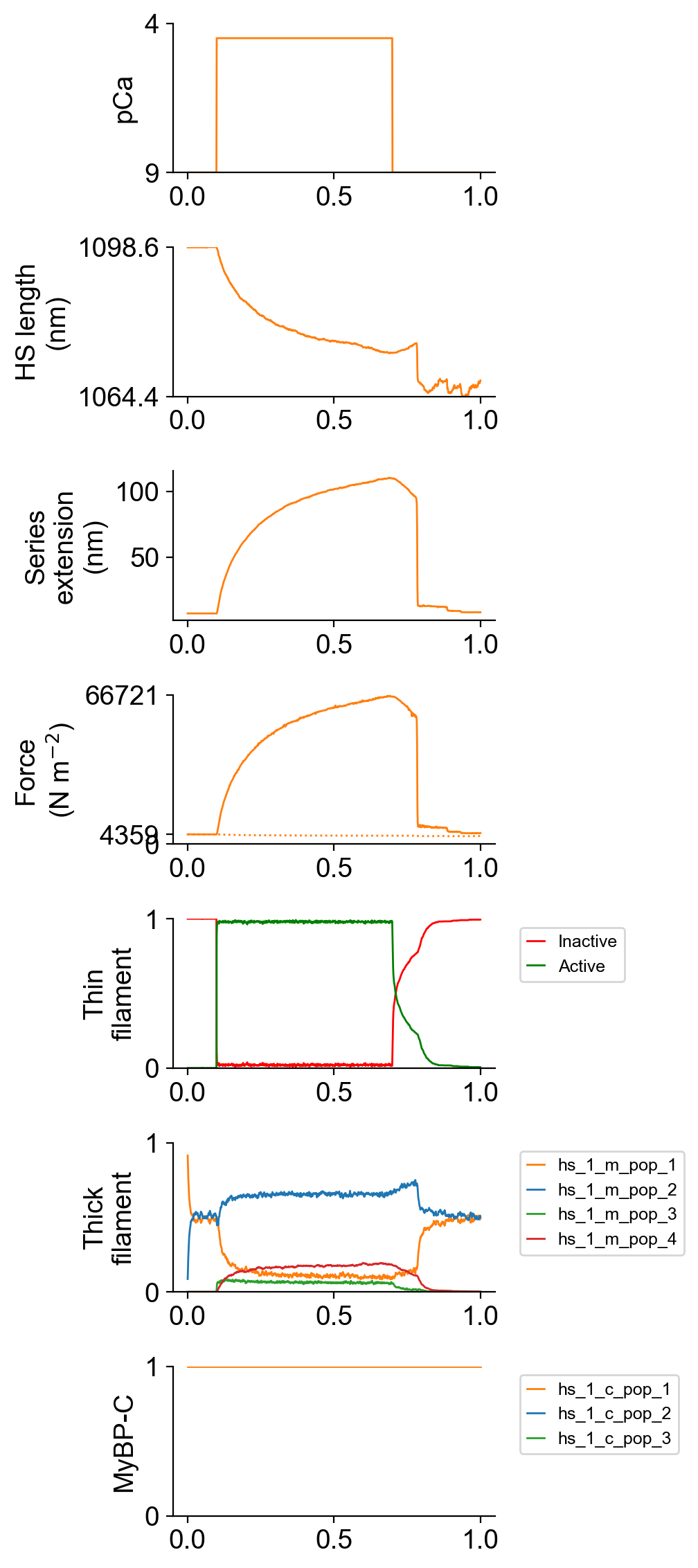
The file superposed_traces.png shows pCa, length, force per cross-sectional area (stress), and thick and thin filament properties for the first half-sarcomere in the myofibril plotted against time. Note the complex time-course of relaxation.
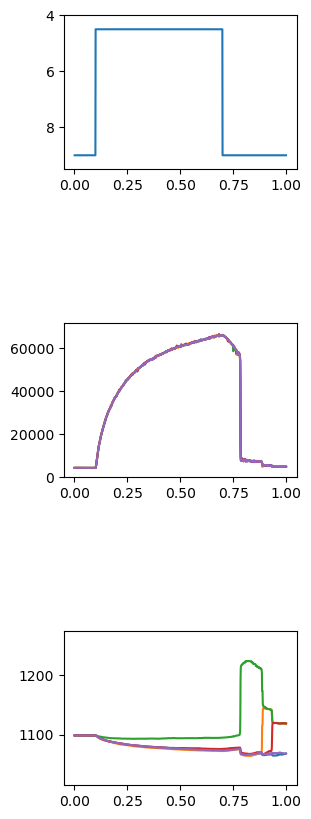
FiberPy also generated a customized figure for this simulation showing the force and length of each of the 5 half-sarcomeres in the myofibril. This file is named summary.png and is saved to the normal sim_output folder.
How this worked
The demo was identical to that shown in multiple half-sarcomeres with series compliance except that the number of half-sarcomeres was reduced to 5 and the rate of the attachment power stroke was reduced in the 3rd half-sarcomere in the myofibril.
The top of the model file (found at <repo>/demo_files/myofibrils/variable_hs/base/model.json) was:
"muscle": {
"no_of_half_sarcomeres": 5,
"no_of_myofibrils": 1,
"sc_k_stiff": 3000,
"initial_hs_length": 1100,
"prop_fibrosis": 0.0,
"prop_myofilaments": 0.5,
"m_filament_density": 0.407e15
},
"half_sarcomere_variation":
[
{
"variable": "m_kinetics_isotype_1_state_2_transition_2_parameter_1",
"multiplier": [1, 1, 0.6, 1, 1]
}
]
Note that no_of_half_sarcomeres was set to 5.
A new section titled half_sarcomere_variation defined an array of manipulations. In this example, the variable name points to:
- myosin isotype 1
- state 2
- transition 2
- parameter 1
The m_kinetic scheme is defined further down the model file. The keys above correspond to a parameter value of 200.
"m_kinetics": [
{
"no_of_states": 4,
"max_no_of_transitions": 2,
"scheme": [
{
"number": 1,
"type": "S",
"extension": 0,
"transition": [
{
"new_state": 2,
"rate_type": "force_and_mybpc_dependent",
"rate_parameters": [ 20, 200, 1, 1, 0.01, 0.01, 1, 1]
}
]
},
{
"number": 2,
"type": "D",
"extension": 0,
"transition": [
{
"new_state": 1,
"rate_type": "constant",
"rate_parameters": [ 100 ]
},
{
"new_state": 3,
"rate_type": "gaussian_hsl",
"rate_parameters": [ 200 ]
}
]
},
{
"number": 3,
"type": "A",
"extension": 0.0,
"transition": [
{
"new_state": 2,
"rate_type": "poly",
"rate_parameters": [ 500, 10, 4]
},
{
"new_state": 4,
"rate_type": "constant",
"rate_parameters": [ 75 ]
}
]
},
{
"number": 4,
"type": "A",
"extension": 5.0,
"transition": [
{
"new_state": 2,
"rate_type": "exp_wall",
"rate_parameters": [ 75, 1, 8, 7]
}
]
}
]
FiberCpp sets the value of this parameter in each half-sarcomere to the base value (200) multiplied by the corresponding value in the multiplier array. Thus:
- half-sarcomere 1, 1 * 200
- half-sarcomere 2, 1 * 200 = 200
- half-sarcomere 3, 0.6 * 200 = 120
- half-sarcomere 4, 1 * 200 = 200
- half-sarcomere 5, 1 * 200 = 200